All content on this site is intended for healthcare professionals only. By acknowledging this message and accessing the information on this website you are confirming that you are a Healthcare Professional.
The gvhd Hub website uses a third-party service provided by Google that dynamically translates web content. Translations are machine generated, so may not be an exact or complete translation, and the gvhd Hub cannot guarantee the accuracy of translated content. The gvhd and its employees will not be liable for any direct, indirect, or consequential damages (even if foreseeable) resulting from use of the Google Translate feature. For further support with Google Translate, visit Google Translate Help.
The GvHD Hub is an independent medical education platform, sponsored by Medac and supported through grants from Sanofi and Therakos. Funders are allowed no direct influence on our content. The levels of sponsorship listed are reflective of the amount of funding given. View funders.
Now you can support HCPs in making informed decisions for their patients
Your contribution helps us continuously deliver expertly curated content to HCPs worldwide. You will also have the opportunity to make a content suggestion for consideration and receive updates on the impact contributions are making to our content.
Find out more
Create an account and access these new features:
Bookmark content to read later
Select your specific areas of interest
View GvHD content recommended for you
Review of oral and gut microbiome signatures in allo-HSCT-associated aGvHD
Hematopoietic stem cell transplantation (HSCT) can induce cure in many acute leukemias but may be associated with acute graft-versus-host disease (aGvHD) which comes with longer hospital stays, increased medical costs, and individuals with Grade 3–4 aGvHD only have a 40% survival rate.1 HSCT conditioning and antibiotic treatment can damage the gut mucosa and create an imbalance in gut microbiota. It is believed that this imbalance and mucosa damage may lead to an increased immune response to the graft, causing aGvHD.1
Micaela Beckman and team, in their systematic review published in Supportive Care in Cancer, using conventional literature search and computational approaches, provide an overview of the oral and gut bacteria associated with aGvHD following allogeneic HSCT (allo-HSCT), and how bacterial metabolic pathways might be involved in protecting from aGvHD.1
Methods
Beckman and colleagues used a combination of methods to identify studies of microbiome profiling with or without aGvHD (traditional search methods) and bacterial taxa associated with aGvHD, and drug interaction analysis (text-mining methods). The methodology used is detailed in Figure 1.
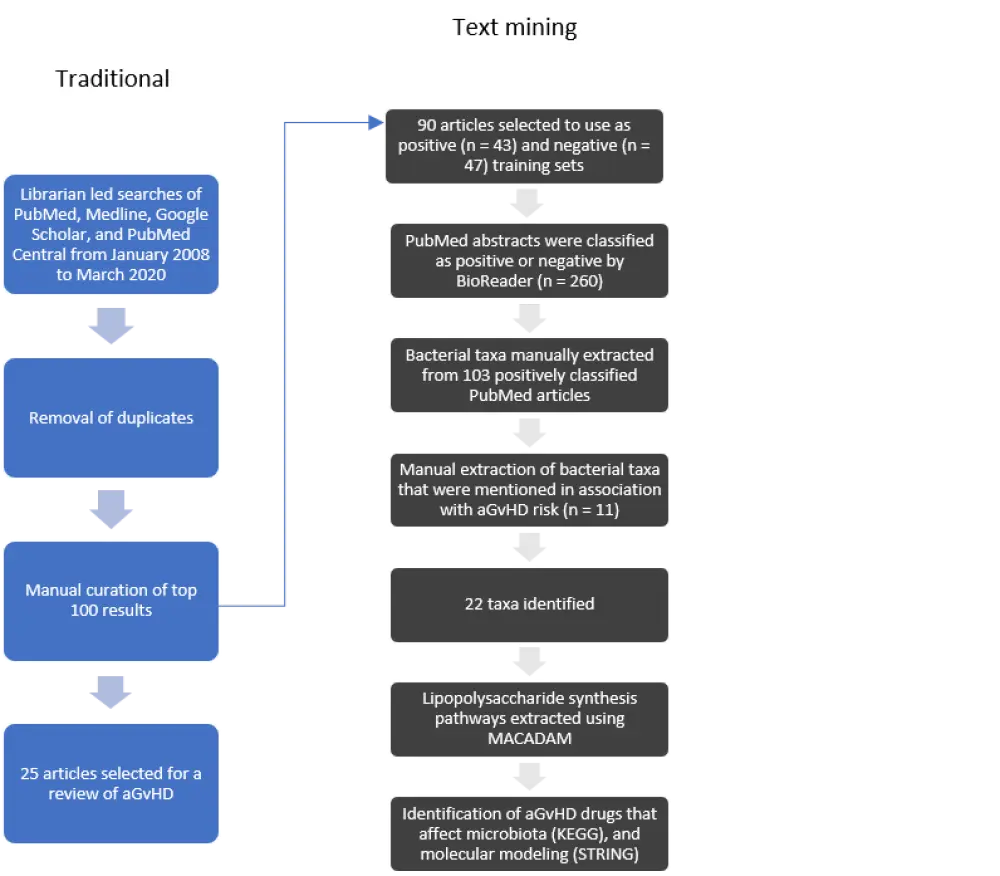
Results
The team found several oral bacteria to be positively associated with aGvHD, and gut bacterial taxa to be both positively and negatively associated with aGvHD (Table 1), with a relative abundance (RA) gradient of bacteria in stools being indicative of severe aGvHD. Several oral genera, Rothia, Solobacterium, and Veillonella, were identified in the stool of HSCT patients and associated with aGvHD. The team found that a change in the microbiome post allo-HSCT was associated with gut aGvHD, and that this was particularly apparent in patients receiving antibiotics for neutropenic infections. Indeed, several of the studies found that dysbiosis of the gut microbiome preceding or during allo-HSCT was associated with a higher risk of aGvHD or a higher morbidity, and that a lower gut microbial diversity in the recipients was associated with higher aGvHD incidence. In studies of pediatric patients, Beckman and team found an association of aGvHD with an RA increase in fecal Enterococcus and Clostridiales, and an RA decrease in fecal Faecalibacterium and Ruminococcus. There was also an association between a higher RA of pathogenic-associated bacteria, such as Staphylococcus and Streptococcus, and an inability to maintain stability of both the oral and gut microbiomes, whereas Akkermansia, Subdoligranulum, and Pseudobutyrivibrio were found to be linked to an increase in stability of microbiome diversity.
Table 1. Oral and gut bacterial taxa identified as potentially associated with aGvHD1
|
Phylum |
Family/ Genus/ Species |
|
aGvHD, acute graft-versus-host disease. *Genera with species identified as being associated with aGvHD. Oral bacteria were identified from stool samples. aGvHD positively (+) and negatively (−) correlated bacterial taxa. |
|
|
Oral microbiome |
|
|
Actinobacteria |
Micrococcaceae/Rothia/ R. mucilaginosa (+) |
|
Firmicutes |
Erysipelotrichidae/Solobacterium/ S. moorei (+) Veillonellaceae/Veillonella/ V. parvula (+) |
|
Gut microbiome |
|
|
Actinobacteria |
Erysipelotrichidae/Solobacterium/ S. moorei (+) Veillonellaceae/Veillonella/ V. parvula (+) |
|
Bacteroidetes |
*Bacteroidaceae/Bacteroides/ B. dorei (+), B. thetaiotaomicron (−), B. ovatus (−), B. caccae (−) |
|
Firmicutes |
Clostridiaceae/Butyricicoccus (−) Enterococcaceae/Enterococcus (+) *Erysipelotrichaceae/Erysipelatoclostridium (−), and Solobacterium/ S. moorei (+) Lachnospiraceae/Lachnoclostridium (−) and Blautia/ B. luti (−) Peptostreptococcaceae (−) Ruminococcaceae (−) *Veillonellaceae/Veillonella/ V. parvula (+) |
|
Fusobacteria |
Fusobacteriaceae/Fusobacterium (+) |
|
Proteobacteria |
Enterobacteriaceae (+) |
Lipopolysaccharide (LPS) is a bacterial surface protein that can bind to the toll-like receptor 4 (TLR4), leading to activation of the NFkB pathway and release of proinflammatory cytokines. In a small pilot study, higher levels of oral Capnocytophaga spp. were found in allo-HSCT patients who did not develop aGvHD, which was believed to be due to the production of a specific LPS that inhibits TLR4. Using the metabolic pathways database for microbial taxonomic groups (MACADAM), the team found that phyla previously identified as positively associated with GvHD using BioReader mostly contained metabolic pathways with links to LPS biosynthesis. When looking at the target genes of drugs used in the treatment and prevention of GvHD, STRING networks highlighted the molecular pathways around TLR4, signaling as drug targets. According to the authors, there are currently no clinical trials on GvHD targeting TLR4. However, an anti-TLR4 drug has been recently assessed for rheumatoid arthritis, which was able to reduce symptoms in more than half of patients.
Conclusion
Beckman and team highlight the importance of the oral and gut microbiomes in aGvHD development, with dysbiosis of the microbiota before or after HSCT being strongly associated with aGvHD. They found that oral bacterial species may, in addition to the gut microbiome, be important factors in the development of aGvHD, and that changes in the diversity of these bacteria need to be better understood in the context of aGvHD. Finally, trials in patients with GvHD targeting TLR4 seem warranted given the link between TLR4 inhibition and lower rates of GvHD.
References
Please indicate your level of agreement with the following statements:
The content was clear and easy to understand
The content addressed the learning objectives
The content was relevant to my practice
I will change my clinical practice as a result of this content
Your opinion matters
Which consideration most strongly guides your decision to escalate therapy in SR-aGvHD?

